Atomsk
The Swiss-army knife of atomic simulations

The Swiss-army knife of atomic simulations

Atomsk was born at some point between February and April 2010, so it turns 10 years old in April 2020. It is the occasion to tell its story: how was it born, how it grew, and how other people contributed to it.
During my Master studies at the University of Rennes, I learned Fortran and programmed my first tools to construct atomic systems: graphite and carbon nanotubes. Then during my PhD at the LMP laboratory (today part of P' Institute) in Poitiers, France, I switched to metallic systems with free surfaces to study their mechanical behaviour and dislocation nucleation, using XMD and LAMMPS to perform simulations, and Atomeye for visualization. After that, I worked as a post-doc at Fraunhofer-IWM in Freiburg, Germany, to study dislocations in perovskite materials, and for that I relied on VASP and LAMMPS for the simulations, and ddplot and VESTA for analysis.
At this point (that was in 2009), I had the feeling I was spending a lot of time (wasting time, really) developping and re-developping codes that were doing the same basic operations: create supercells, convert files from one format to another, and so on. I had different little programs to do all that, which contained a lot of duplicated code, and were hard to maintain or to expand. Around me, many colleagues also had their own custom codes, specific to their research goals and to the type of simulation or analysis they were performing. Nobody was sharing their codes, because they were too specialized, hard to expand, and let's be honest, for the most of them, developped in haste, buggy and in many different languages.
That's when I decided to create a program to create, manipulate and convert atomic data files, that would be general and easy to expand. One program that would support all types of atomic structures, no matter the lattice (fcc, bcc, perovskite...) or the chemical composition. One program that would support all the different file formats that I was using, so I could easily convert from one format into any other (LAMMPS, VASP, Atomeye...). One program that would implement all the elementary operations that everybody needs: create a unit cell, orient it, duplicate it, and so on.
I had to give that program a name, and since it was not meant to be shared I called it "PIKASHU", a funny acronym that meant "PIerre's Swiss-army Knife for handling Atomic file formats in a Simple and HUman way". A version was labelled "Beta 0.1" on April 27, 2010 -let's consider this date as the date of birth of this program. It was really just a patchwork of the different routines I had developped through the years, each of them reading an XYZ file and writing another file. That required a lot of disk input/output every time an operation was performed, which was highly unefficient. Below is a version that dates from June 16, 2010, one of the earliest versions that I have retrieved on my computer.
As the only user, I was also the most critic about this version. At my workplace, all users' directories were remote, and it took a painful amount of time every time I had to run this code to convert files. By the end of 2010 I removed all this useless disk I/O, and all the data was passed between the routines -everything happened in the computer's memory (RAM), making the program much faster.
PIKASHU attracted the curiosity of collegues, who soon asked me to share it. Matous Mrovec, Adham Hashibon, Pavel Marton, and PhD students at Fraunhofer-IWM were among the early users of the program. They reported bugs and made suggestions that improved the code in many ways.
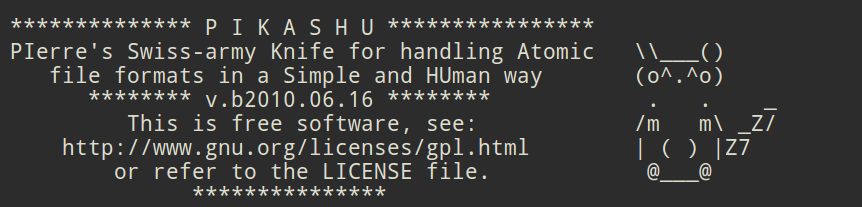
The header that early users saw in their terminal every time they ran PIKASHU.
In order to keep track of the changes I started using a versioning software, and at first I chose SVN which I ran on my own private server. Releasing my buggy code to other people also made me think about the license. From the very start, I decided to make it Open Source under the General Public License, because I believe that a scientist should share his or her receipes, and not hide behind a "black box" program. Everybody should be able to see how I implemented things, and point out any mistakes that I had made. That could only help me fix the errors and improve the program.
In 2011 Matous Mrovec helped me implement the resolution of the equations giving the displacements due to a linear dislocation in an anisotropic medium. I started from one of his own codes (with some comments written in Czech), which I adapted and plugged into Atomsk. Solving these equations introduced the dependency to the LAPACK library. Around that time, I also decided to make PIKASHU publicly available on my personal page.
Soon after starting a new post-doc at UMET laboratory at University of Lille, France, I made several decisions concerning this code. First I wanted to change the name into something that would be more simple to remember and related to atoms and atomic-scale simulations: in 2013 it became Atomsk. One year later I changed the versioning method to GIT and a public GitHub repository to share its developments. I did not care to convert the older SVN history into GIT, so the version history previous to the initial GIT commit in 2014 was simply lost.
The code continued to grow in size and functionality. By 2015, Atomsk supported auxiliary properties, shell-model potentials, allowed to merge atomic systems, construct polycrystals, and many other features. Philippe Carrez implemented a module for computing the Nye tensor. I decided the code was mature enough to write an article to promote it, and it was published in 2015 in Computer Physics Communications.

The same year I also decided to create a Web site dedicated to Atomsk, instead of sharing it on my personal page. The University of Lille provided the hosting facilities, and since 2015 Atomsk has its official home page offering tutorials, the documentation, the source code and binaries to ease the installation process.
This new availability and visibility attracted new users, who soon started to report bugs and suggest program improvements. One large improvement was performed by Juri Bartel, developer of the TEM image simulation software Dr Probe: he implemented the space group symmetries in Atomsk, and improved the support of Crystallographic Information files (CIF). This big contribution allowed me to extend the support of space groups in other file formats as well (GULP, VESTA, etc.).
In 2018 I contacted Emmanuel Clouet, the developer of the Babel package, to ask for his permission to include his routine for generating dislocation loops. Being the nice person that he is he agreed, and now that feature is also part of Atomsk.
Thanks to all these developments and contributions, Atomsk is now used by people all around the world, as evidenced by the list of articles citing the Atomsk paper grows every month. As it turns 10 years old, Atomsk has been cited in 143 scientific journal articles.
At the moment when these lines are written, the version available from the Web site is labelled 0.10.6, which was released in October 2018. That was a long time ago. In the mean time, many bugs were corrected, and many features were added: parallelization with OpenMP, support for ABINIT, BOPfox, CRYSTAL and SIESTA formats... All these modifications are freely available from the GitHub repository, but I did not have time to prepare a new release. I intend to do so at some point during 2020.
As you can see from the download page, Atomsk is still in "Beta version". Indeed, I consider that it is still "work in progress", and I know that there must be some bugs lurking in the code. I encourage all users to report bugs if they see any, in order to improve the robustness and reliability of the code.
Happy birthday to Atomsk! I want to thank all users and contributors for their work and their efforts, for using Atomsk, and for making it a better program. Stay tuned for more adventures!